SHORT NEWS
Breakthrough in protein recognition - thanks to Deep Learning
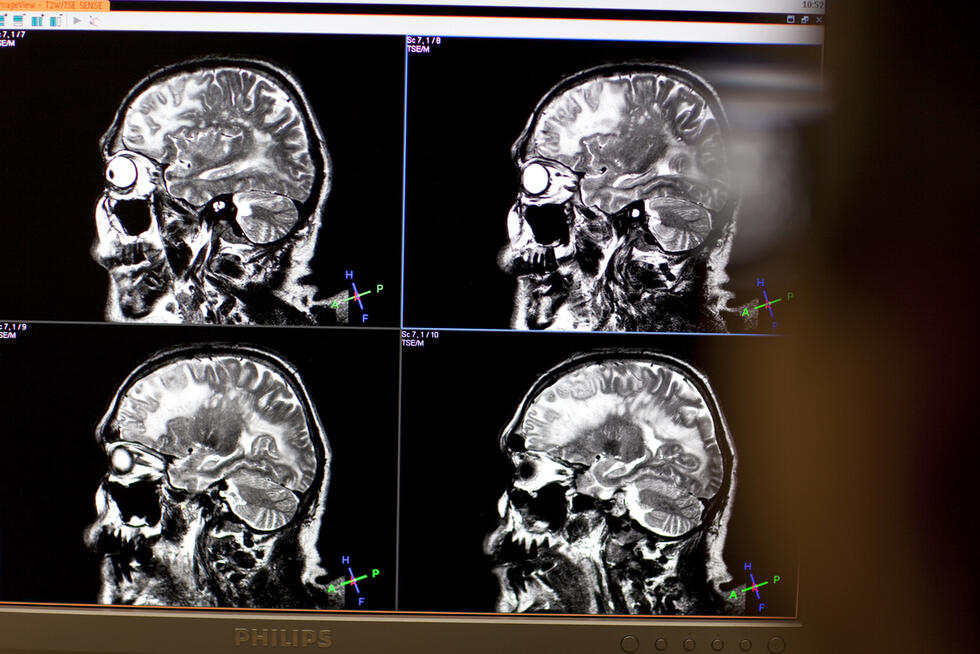
Proteins control most processes in living cells and are vital for humans. If it were possible to precisely determine all proteins in an organism, diseases such as Alzheimer's or cancer could be treated better and more individually.

Scientists at the Hasso Plattner Institute (HPI), led by Professor Bernhard Renard, together with other partners, have now succeeded in coming closer to this goal: by combining a deep learning model with conventional algorithmic methods. In the renowned scientific journal Nature Machine Intelligence, they present "Ad hoc learning of fragmentation" (AHLF), a deep-learning model that was trained on mass spectra with almost 20 million data points and was able to improve phosphopeptide identifications by up to 15 percent compared to conventional methods.
In addition to the more accurate analysis and evaluation of proteins, the interpretability of AHLF is another advantage. "Compared to many other approaches, AHLF is not a black box, but interpretable. It is therefore transparent how accurately AHLF recognises patterns of proteins in the mass spectra, some of which have been less researched or not yet researched at all," says Professor Bernhard Renard, who heads the Data Analytics and Computational Statistics department at HPI.